Warfarin Design
Contents |
Background
Warfarin predictor is a result of my cosc366 research project. It is a web based application that uses machine learning algorithms to predict a right dose for heart-valve transplant patients.
What is Warfarin?
Warfarin is taken by patients as a blood thinner in order to prevent blood clotting In It is very important that right dose of Warfarin has been taken so that patient has a minimal chance of clotting, while still ensuring that patient has enough clotting ability so that he or she does not bleed to death. Blood tests needs to be performed in order to know if INR (International Normalised Range, a normalised measure of the time taken for blood to clot) reading for the patient is within a safe range. The target INR range differs depending on the type of valve replacement, and to some degree and also on the patient. The way the patient will react to Warfarin intake depends on many factors. These include an individual's age, weight and gender, lifestyle habits such as alcohol and tobacco consumption, and even environmental factors. So how to help doctors prescribe the right dose of Warfarin? It is hoped that modern technology particularly a machine learning solution could be the way to go. So this is where my journey starts.
Project goal
The main goal of this research is to build on what is learned by in now in domain of using a machine learning to prescribe Warfarin, and develop a prototype for web-based Warfarin Predictor. This prototype will need to allow for a doctor to have enough flexibility to create and combine different attributes (information that a doctor would find valuable) in order to create a model that doctor can then use to predict Warfarin intake of the patient with the greatest accuracy. By giving doctor enough flexibility to self create the input attributes and combine them in order to make a prediction model. It is hoped that it would be possible to find the combination of attributes and algorithm that could best suit individual patient. This could be particularly important when the patient does not have a long history of taking Warfarin and other patient’s history is to be used. The web-based prototype is not going to give one best solution for all it would act more as a tool that is going to be used by doctors to help them make a right decision. It is hoped it may improve accuracy of dosage, cut costs, decrease physician workload and help with keeping an accurate patient medical history. The other goal of this research is to do some experimental work once the web-based predictor is created to give some insight to what type of attributes should work best with which machine learning algorithm as it is expected that some combination would not give any value while others could lift the accuracy of Warfarin prediction.
Requirements
Initial Requirements
- System users: administrators, doctors.
- Administrators can add new doctors.
- Doctors adds patients, patient records.
- Main use case is to record a new blood test (INR reading), get a dose prediction.
- Dose predictions may be returned in several ways, probably not advisable to just give the dose to prescribe because of liability issues. For example, could give a graphical representation of the likelihood of being below the safe range, in range or above.
- The system needs to be able to "learn" a model from the historical data. This involves transforming the data into a special format (an "ARFF" file) and submitting this to the learning algorithm.
- The data-to-arff step needs to be flexible (i.e. able to be easily changed if the learner's data requirements change.
- Models will need to be persisted.
- The data needs to be reliably persisted.
Future requirements
- There may be a need to record other data for the patient, possibly temporally, such as diet, other drugs taken, smoking and alcohol consumption.
- Generate graphs from multiple classifiers (predictions coming from different machine algoritm) to present to the doctor
- Investigate other ensemble methods (combine the prediction coming from different machine learning)
- Investigate using more classes (e.g. not just high and low but also very high and very low)
Current system design
Initial design
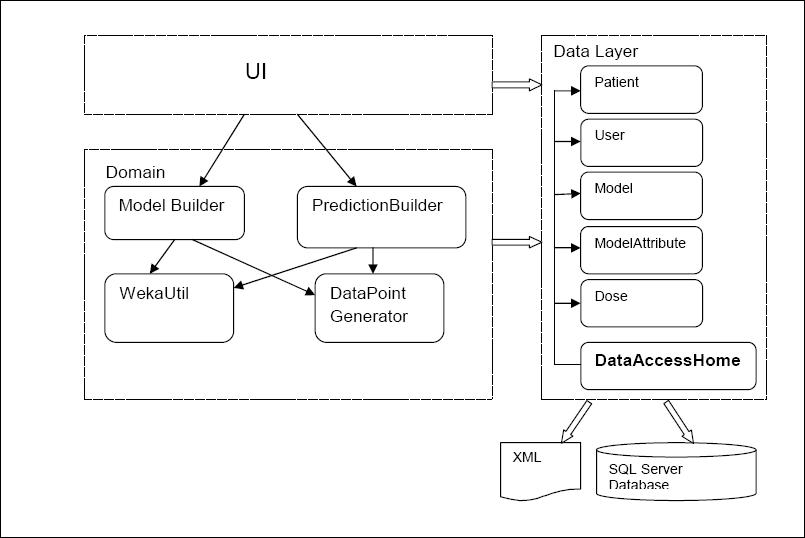
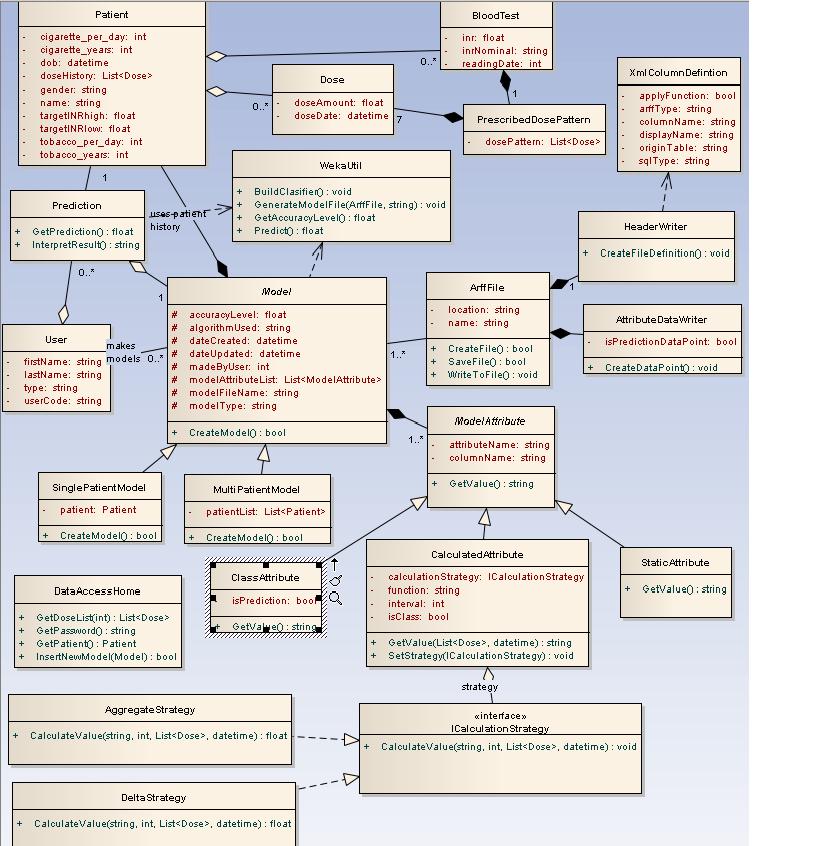
The high level presentation of classes used and their mutual interaction
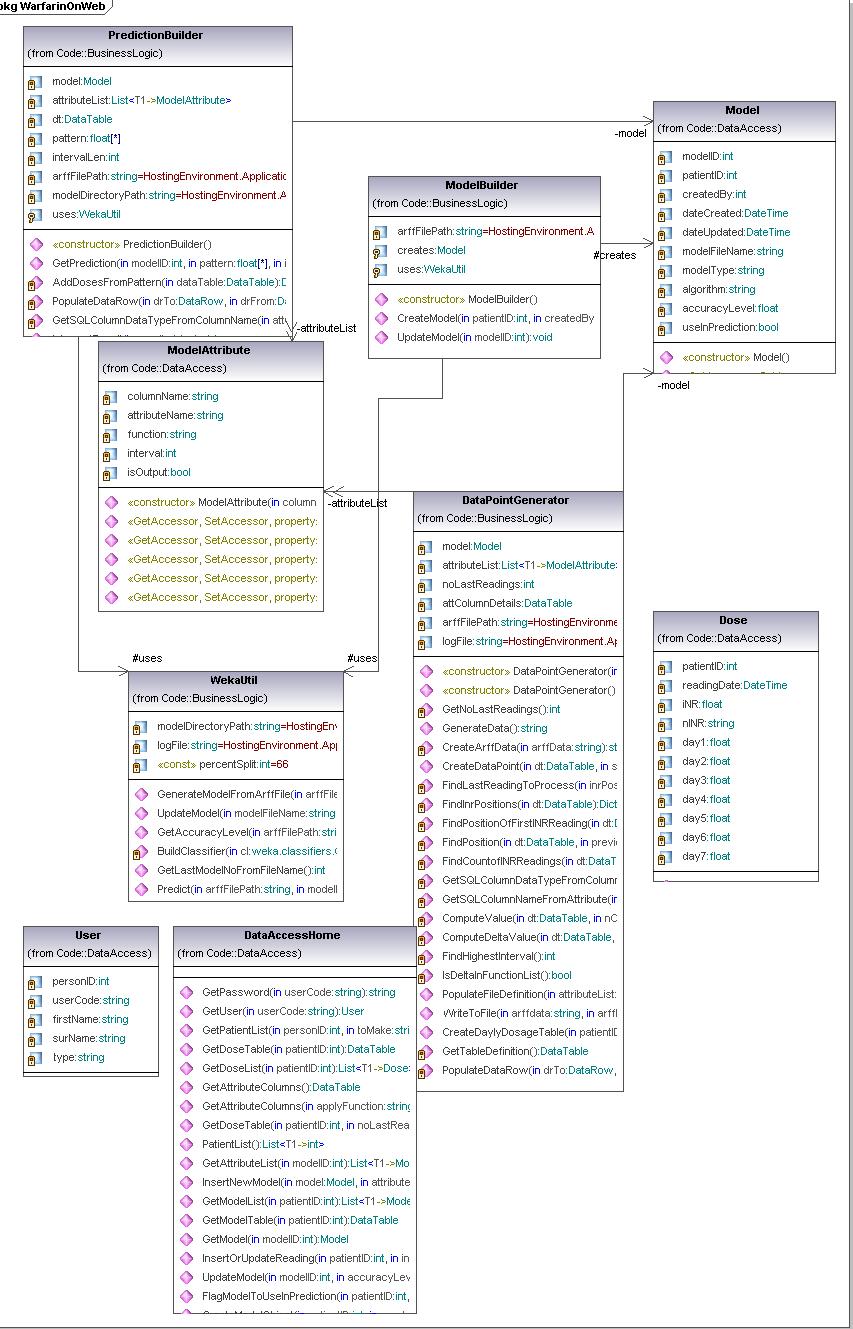
Uml diagram
Class description
Domain logic is made out of 4 classes:
- ModelBuilder wraps up the logic for new model creation and the updates of existing models. It makes appropriate calls to WekaUtil and Datapoint Generators and data source as needed. Once the model is created and updated it invokes the data layer methods for persisting the model to the database.
- PresentationBuilder gives back the INR prediction. It uses DataPoint Generator to make the datapoint , and then it passes it to the WekaUtil to get the prediction for it. It also interprets Weka’s result depending on whether the predicted INR reading needs to be nominal or numeric. It is also responsible for making appropriate calls to data layer to persist (log) completed predictions.
- DataPointGenerator has the datapoint creation logic. It loops through the patient’s dose history and based on the attributes selected it calculates the amounts, comma separates them and puts them in to the datapoint. Its final product is an arff file (made of numerous datapoints) needed for model creation and updates as well as for making a prediction.
- WekaUtil process the API calls to Weka in order to physically create and update a model, persist it to the file, calculate the accuracy level of model created as well make a prediction on the generated datapoint.
Data Layer is primarily responsible for storing persistent data.It is made of one class - DataAccessHome. It communicates with an SQL server database (reading and write data to) and xml file (reading data).The pattern used as a starting point for this layer was Table Data Gateway pattern, where an object acts as a Gateway to a database table. As this pattern suggests there should be an object for each physical table in database where table data Gateaway “holds all the SQL for accessing a single table or view: select, inserts, updates and deletes. Other code calls its methods for all interaction with the database.” (Fowler 2003) This pattern was slightly adapted when designing our prototype due to fact that our problem hasn’t been addressed as big enough to have Table Data Gateway for each table in database, it was considered that one Table Data Gateway(DataAccessHome) would be enough to handle all methods for all table (as suggested for simple apps).
Due to the nature of the problem where we would be able to group all users’ actions into three main streams: create model, update model or making a prediction. Those actions could then be broken down into the many subroutines, where many of those would be shared between those main action points. It then makes a perfect sense to organise our domain logic as suggested in Transaction Script pattern. A Transaction Script is essentially a procedure that takes input from the presentation, processes it with validations and calculations, stores data in the database, and invokes any operations from other systems.(Fowler,2003) So it made a pefrect sence for me to use this pattern at the time.
The way the system currently works
The way the system currently works is as follows:
- User(doctor) logs in.
- From here doctor has two options: It can either
- Create a new model from a patient
- Make a prediction for a patient
- If he chooses to create a new model he needs to
- Select a patient
- Select a type of the model he wants to create (single or multi patient model). If it is a single patient model only the chosen patient dose history would be used to “learn” a model. If the multi patient type is chosen the dose history for all patients would be used.
- Next thing to do would be to select the model attributes that would be used to “learn” the model.When creating an attribute there are a number of different functions that can be applied to any numeric data field , such as sum,average and delta.Each function can be applied to different length time periods.Calculated attributes can be repeated to up to four times per example(for the current and previous three dosage intervals). General attributes (e.g. gender) can also be added or removed from the final group of attributes. Finally, there are potentially many different ML algorithms that can be used to create a model; the prototype uses WEKA code base as an source of ML algorithms and doctor can select which ones they wish to use for a given patient.
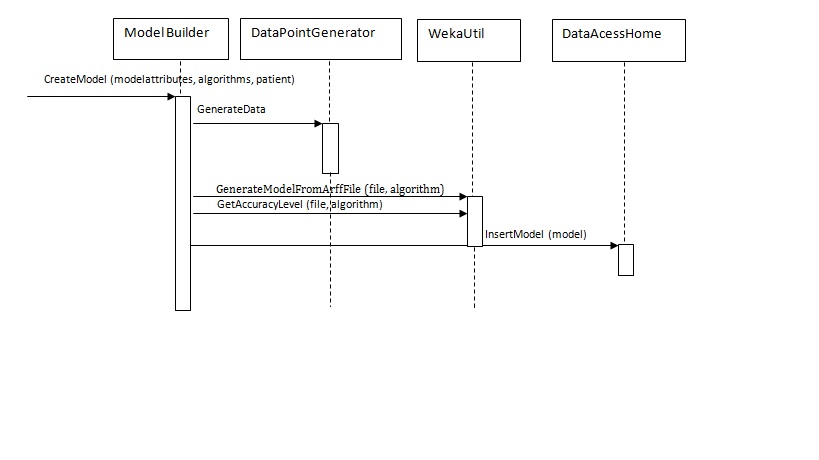
- Once all the above selection are made the user clicks on “CreateModel” button. This would invoke the CreateModel method of the ModelBuilder class passing the list of ModelAtttributes objects created by selecting the attributes from previous screen. The list of chosen algorithms would be passed as well as the patient the model gets created for.CreateModel function would then create DataPointGenerator object and invoke appropriate methods to create the arff file based on patients doseage history and model attributes selected on the previous screen.Once the Arff file gets created CreateModel method would pass it’s location path as well as the selected algorithm names to WekaUtil object. WekaUtil makes appropriate API calls to WEKA’s machine learning algorithms and the model file gets saved on the disk(WekaUtil), and persisted in the database(ModelBuilder).
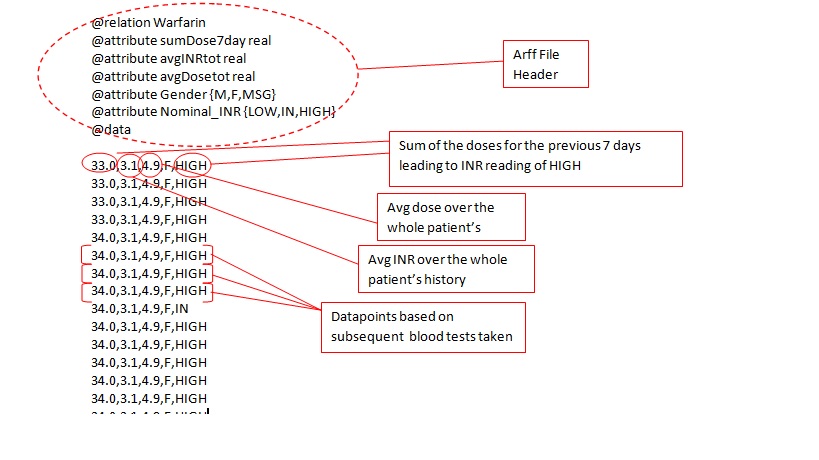
Arff file example:
The sequence diagram for Creating model:
- If the user chooses to make a prediction
- Selects a patient to do prediction for
- Selects a model to use for a prediction (can multi select)
- A doctor needs to enter a new blood test results and also the new week long dose pattern that would like to get INR prediction on. If the doctor doesn't enter the desired pattern every 0.5 mg in the range of 0-15mg is used to predict on.To askive this GetPrediction method on PredictionClass is called from the presentation layer.GetPrediction methods invokes all needed actions in sequance.Starts with the GenerateDataPoint methods to get the Arff file for given dose pattern .Similare to creating a model, the arff file location gets passed to WekaUtil MakePrediction method. Lastly prediction gets persisted by DataAccessHome object.
Critique
- Open/close principle .One of the main problems here for me personally is the lack of flexibility. When creating an arff file (formatted historical data used to "learn" a model) only limited set of functions(hardcoded) could be used to do so(sum,avg,delta,min,max).In the case when we would like to add a ratio as an a function to be used many parts of the code would need to be touched. The computation at the moment is done in DataPointGenerator by using ComputeValue(used for computing data using aggregate functions) and ComputeDeltaValue(used to compute delta value).
- Model the real world The existing design doesn't model the real world. DataPointGenerator does most of the work regarding processing of the data and producing the arff file. ModelBuilder has a create model function that should have been the part of model itself.
- Keep related data and behaviour in one place Data and behaviour are not kept together. Model,ModelAttributes, Patient are only carrying the data and don't have any behaviour.
- Separation of concerns the program is not divided into small, logical, self explanatory classes. The classes either do too much , or not enough.
- Avoid god classes The DataPointGenerator is definitely a god class here. It has about 50% of functionality embedded in itself.
Ideas for improvements
Improved design
Following the smells and basic design principles I have come up with my first version of the improved design. The Uml follows: